Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
Por um escritor misterioso
Descrição

Modelling peptide–protein complexes: docking, simulations and machine learning, QRB Discovery

Shown are chemical structures of the correctly predicted non-substrates
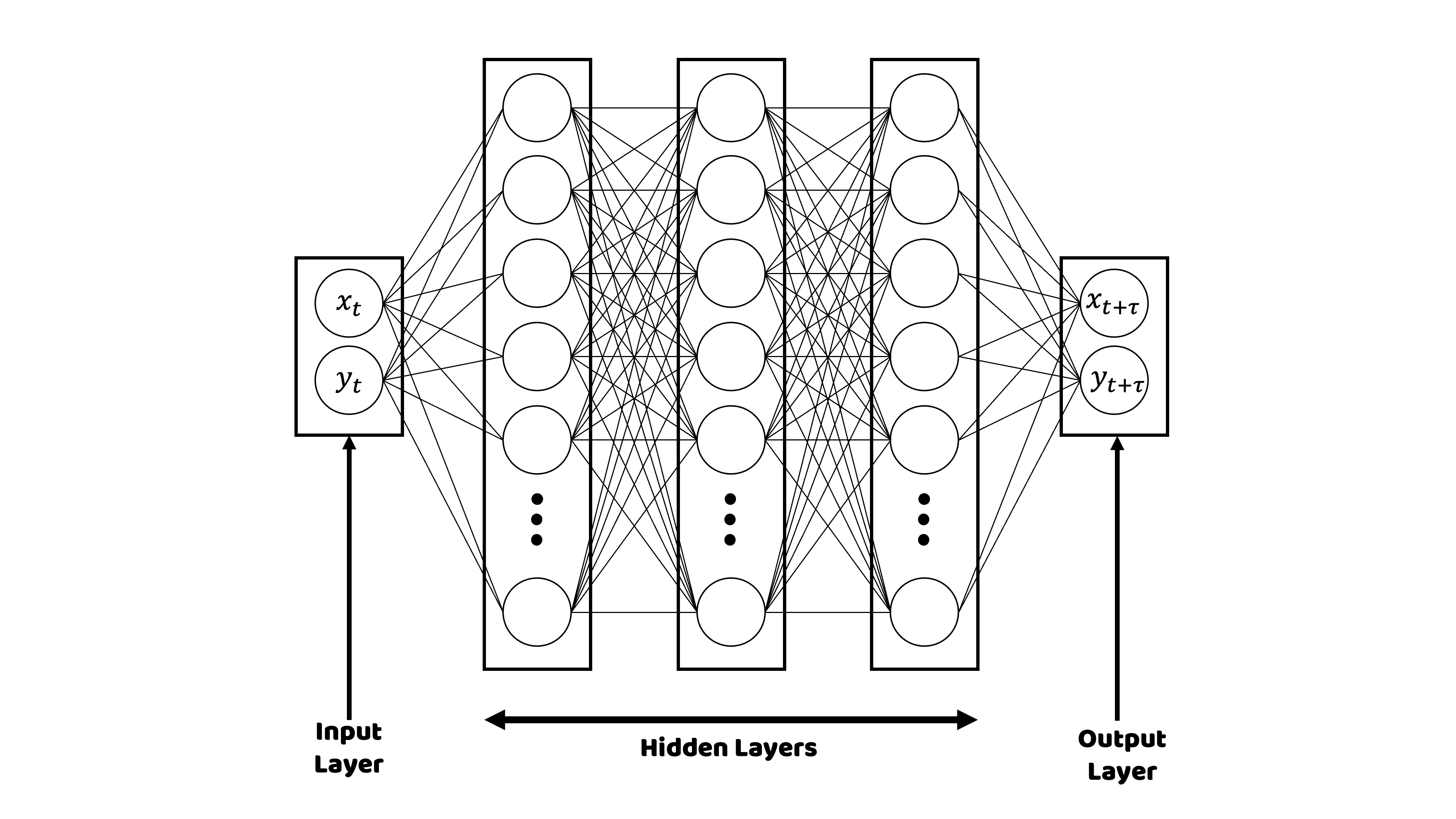
BioSimLab - Research

Structures of P-glycoprotein in different conformations (A) Domain

Development of Simplified in Vitro P-Glycoprotein Substrate Assay and in Silico Prediction Models To Evaluate Transport Potential of P-Glycoprotein
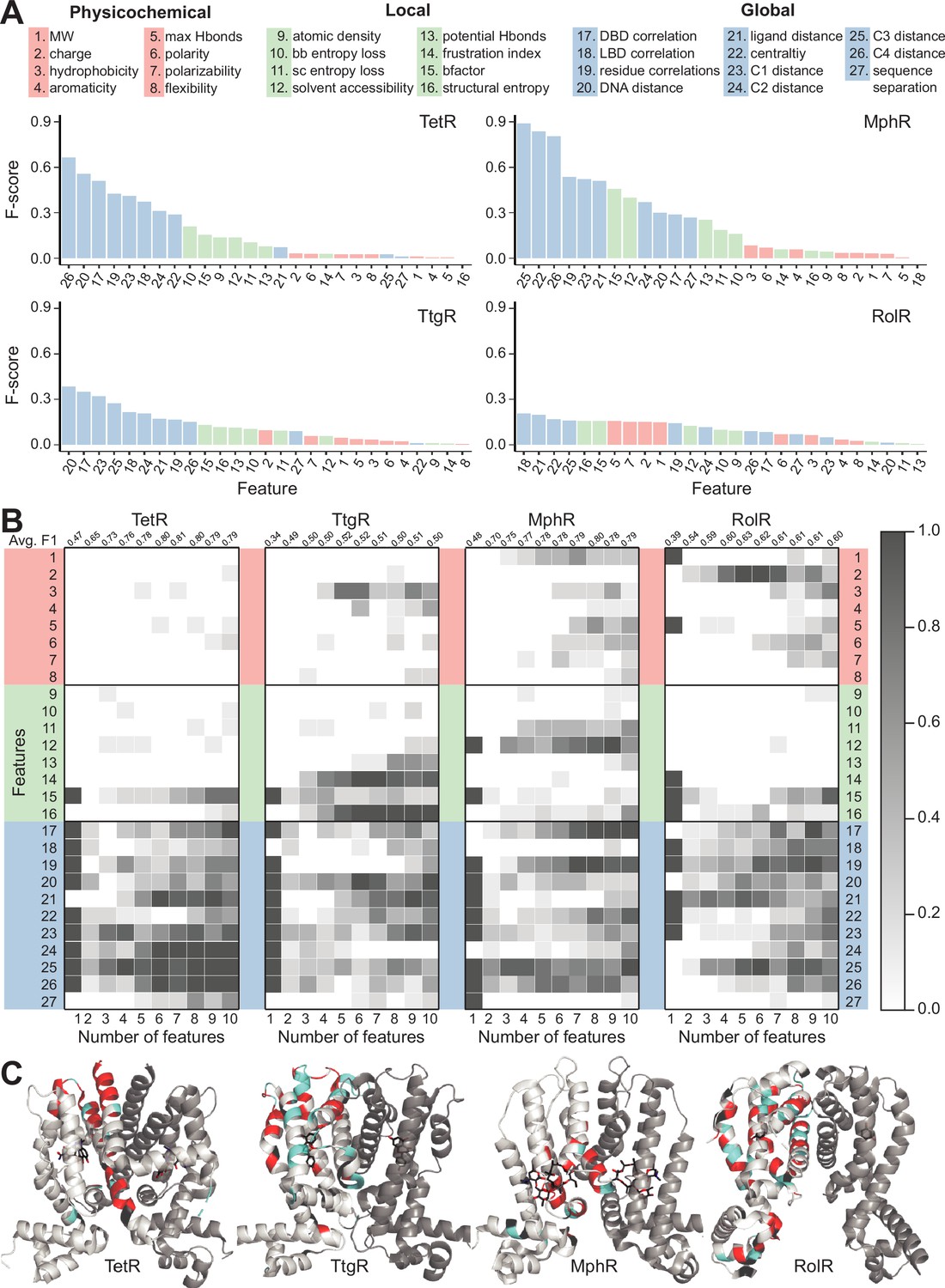
Deep mutational scanning and machine learning reveal structural and molecular rules governing allosteric hotspots in homologous proteins

Computational and artificial intelligence-based approaches for drug metabolism and transport prediction: Trends in Pharmacological Sciences

P-gp substrate probes (A), known P-gp substrates and an MRP2 inhibitor

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates

Machine learning for small molecule drug discovery in academia and industry - ScienceDirect
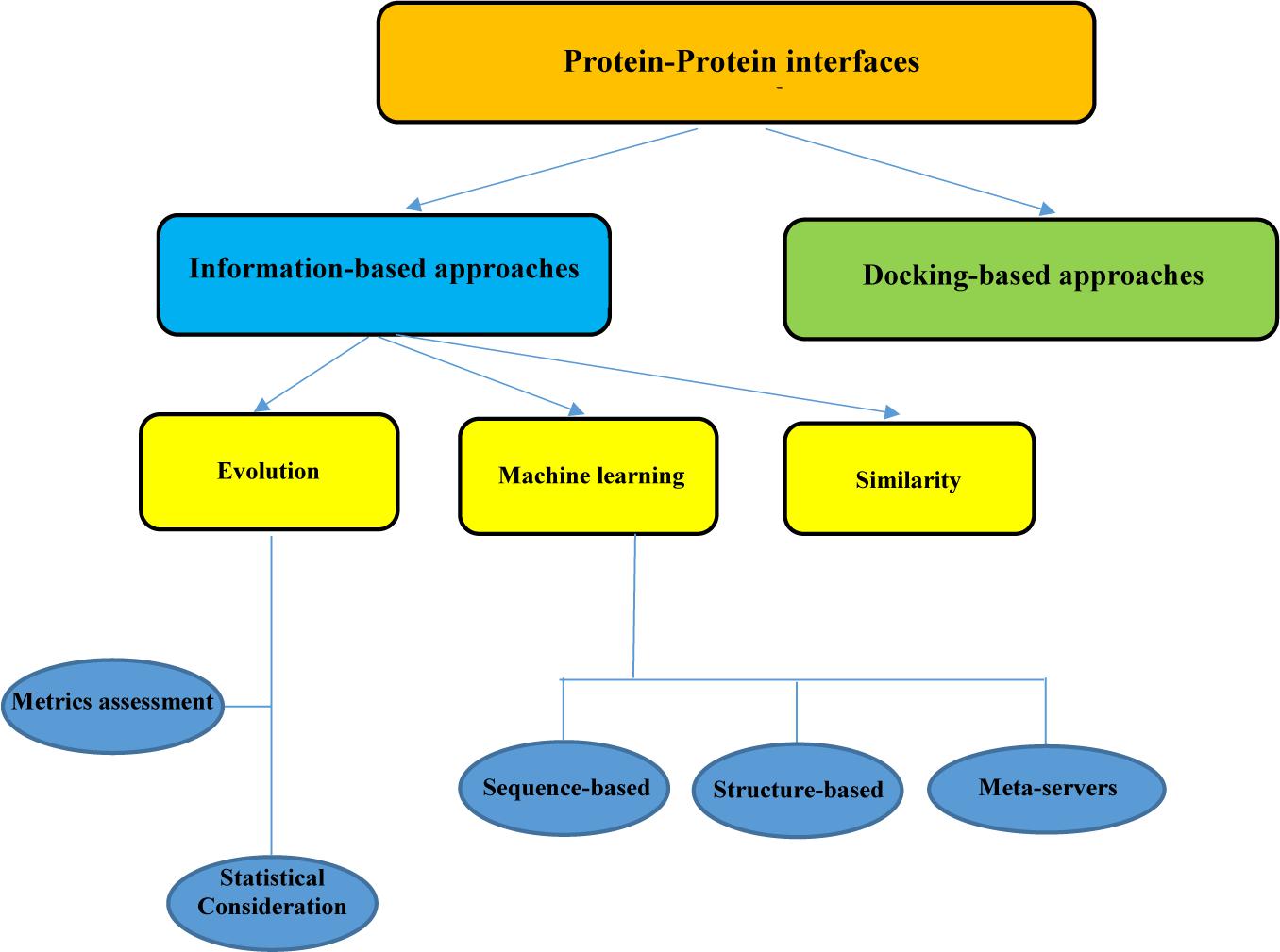
Frontiers In silico Approaches for the Design and Optimization of Interfering Peptides Against Protein–Protein Interactions
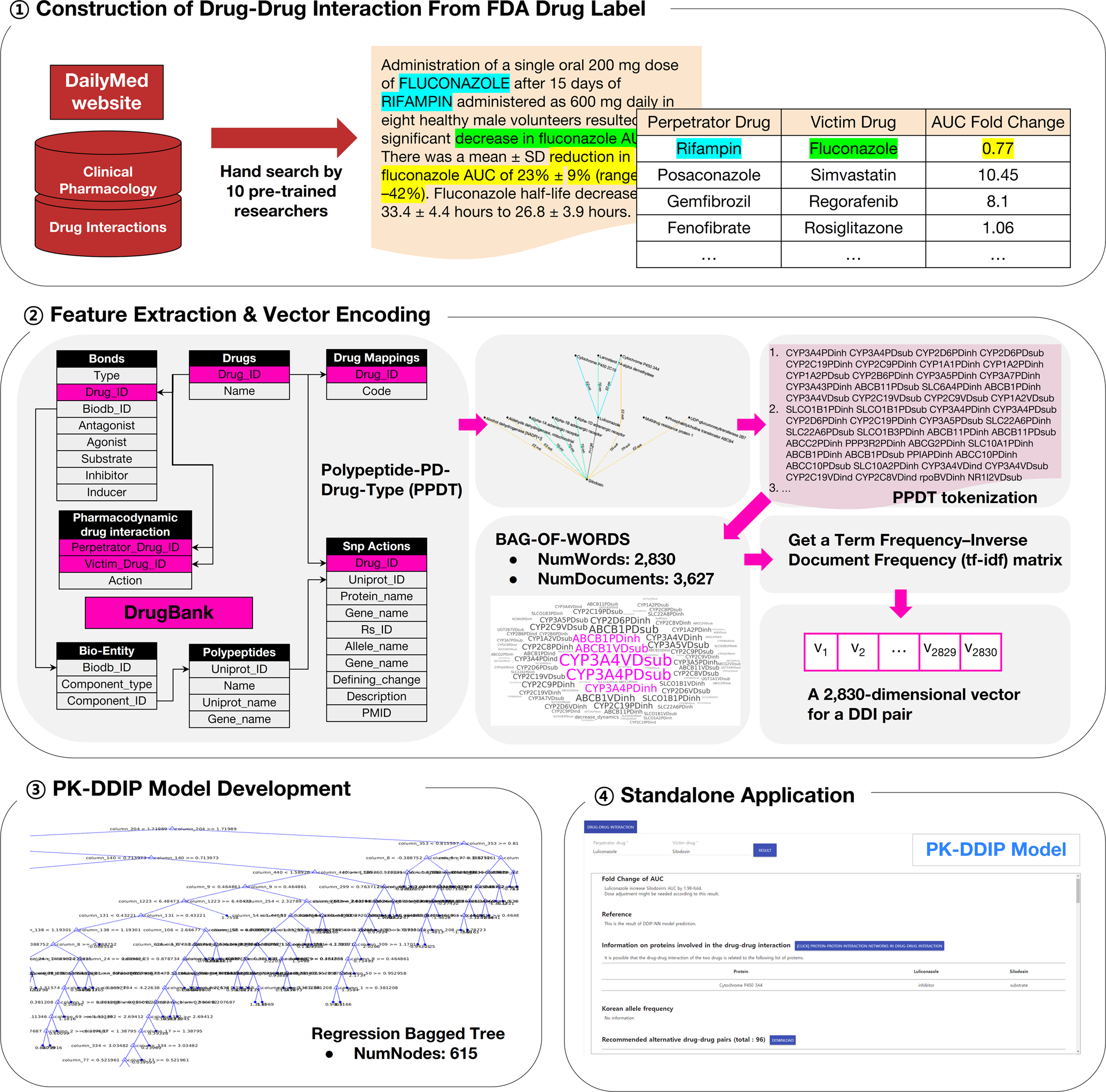
Machine learning-based quantitative prediction of drug exposure in drug-drug interactions using drug label information
de
por adulto (o preço varia de acordo com o tamanho do grupo)







