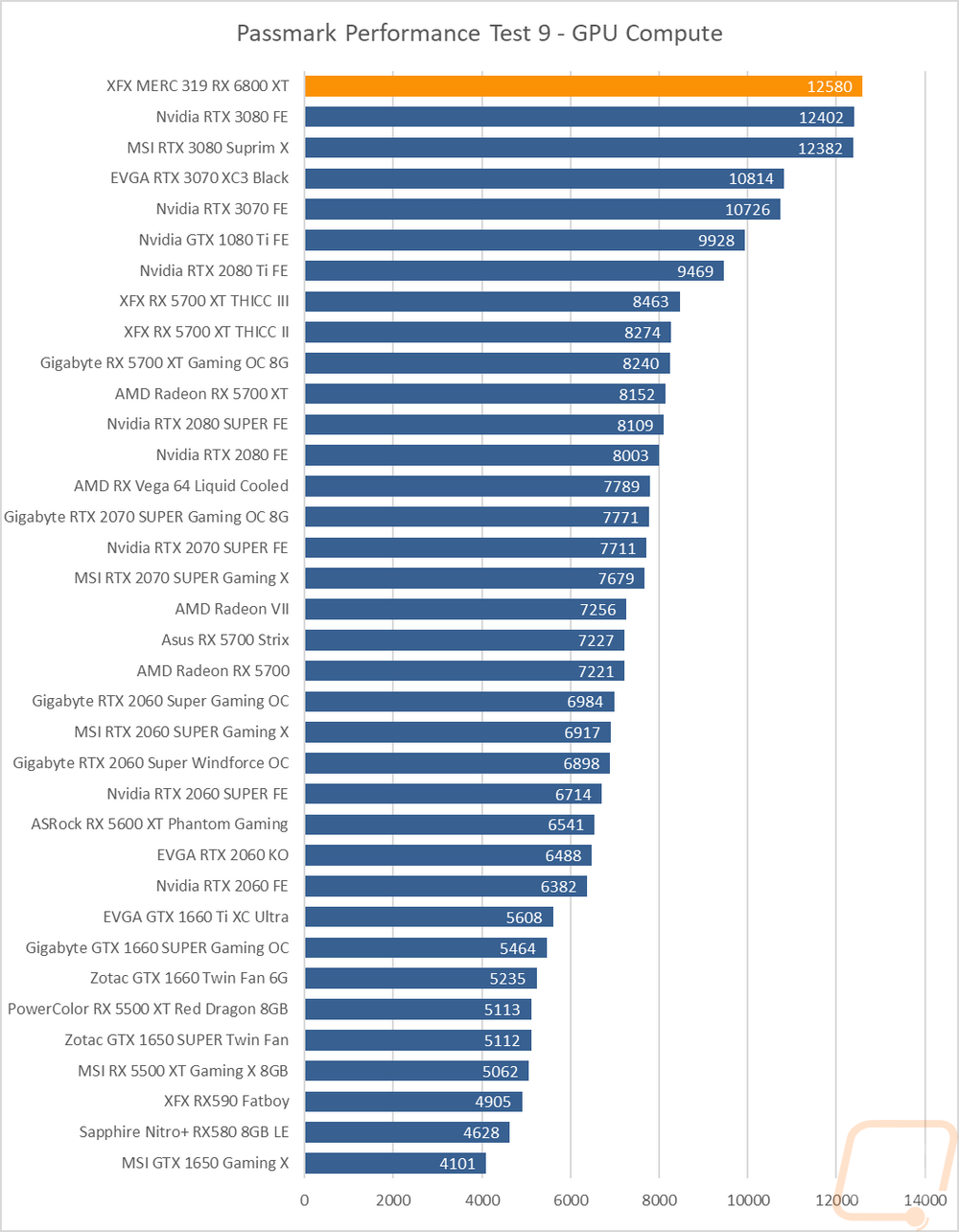
The pmemd.cuda GPU Implementation
Por um escritor misterioso
Descrição
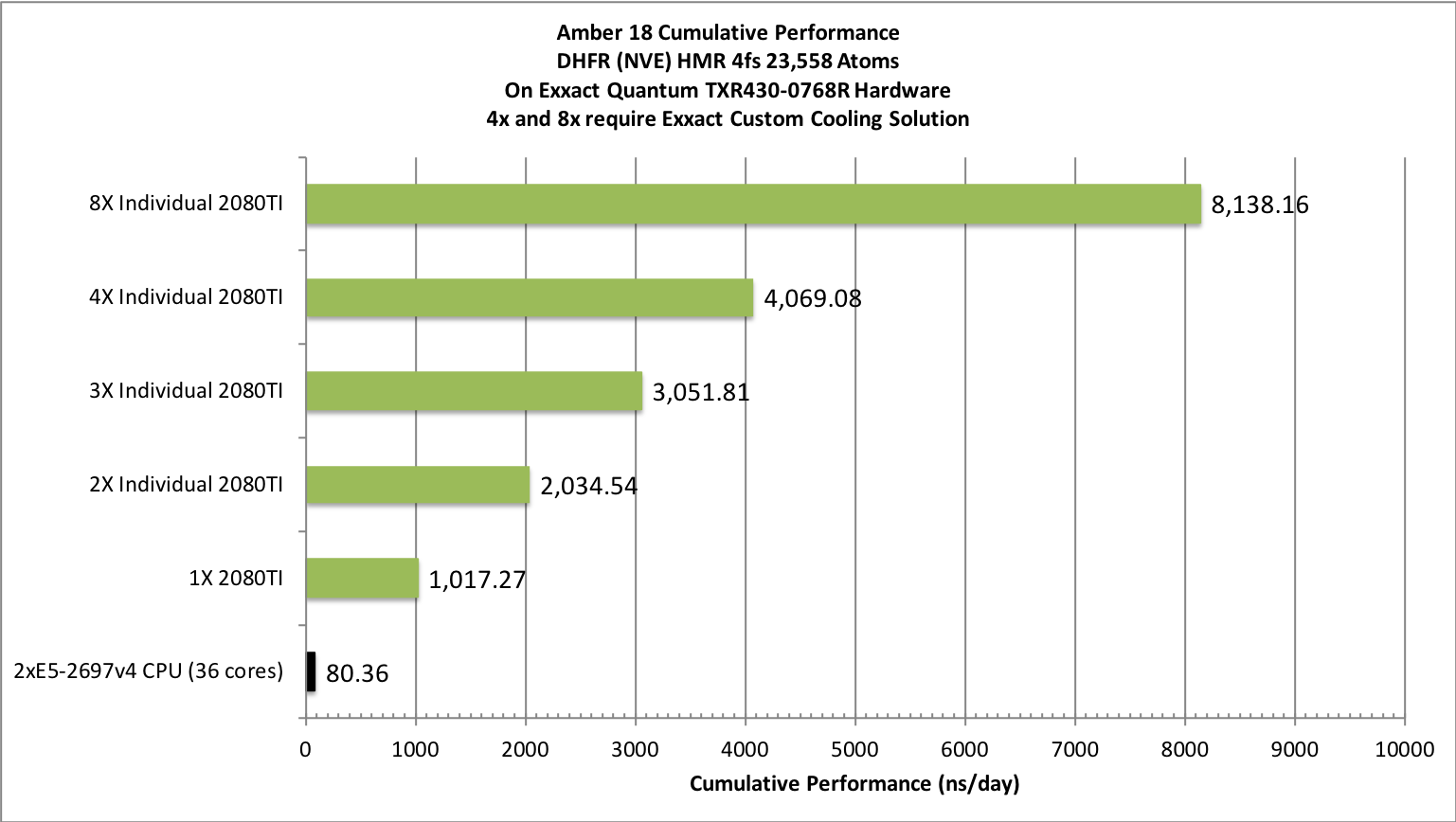
The pmemd.cuda GPU Implementation
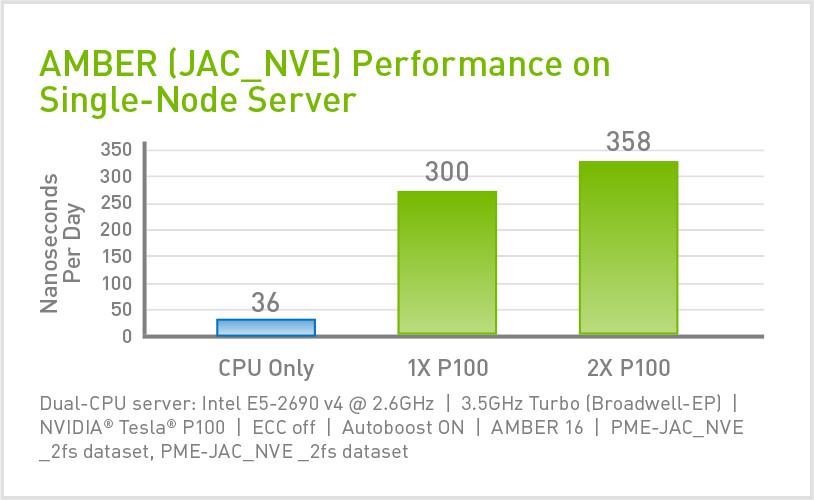
Amber Hardware & Software Configurations

GPU-Accelerated Molecular Dynamics and Free Energy Methods in Amber18: Performance Enhancements and New Features

Performance of Amber 18, NAMD version 2.13 and GROMACS 2018 measured as
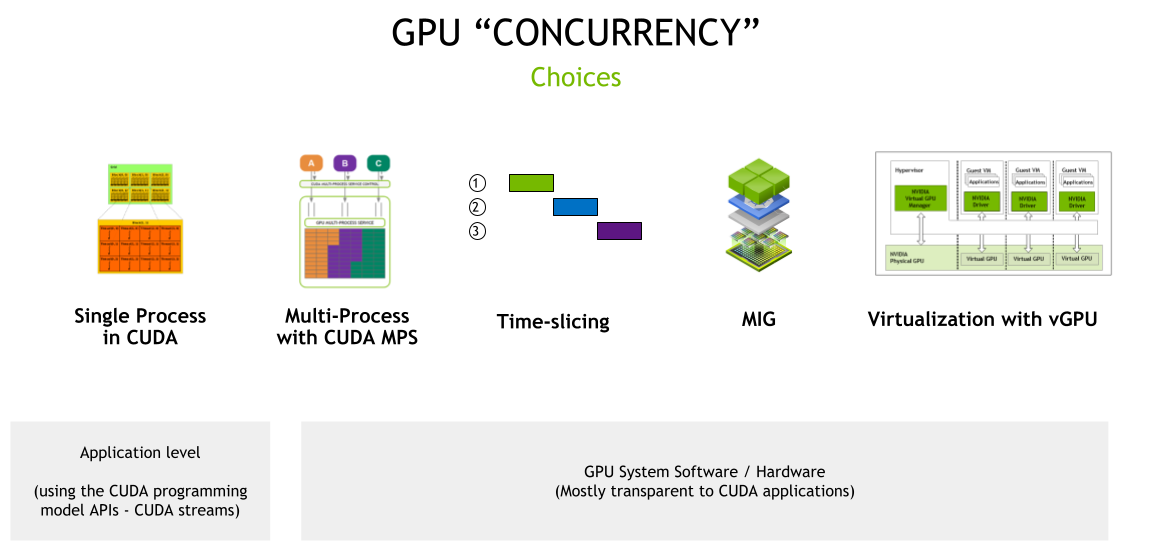
Improving GPU Utilization in Kubernetes

Schematic representation of the main components of the MIST library.

Toward Fast and Accurate Binding Affinity Prediction with pmemdGTI: An Efficient Implementation of GPU-Accelerated Thermodynamic Integration.

GPU-Accelerated All-atom Particle-Mesh Ewald Continuous Constant pH Molecular Dynamics in Amber

GUI and output for statistical comparison of dynamic motion

GPU-Accelerated Implementation of Continuous Constant pH Molecular Dynamics in Amber: pKa Predictions with Single-pH Simulations

GPU-Accelerated All-atom Particle-Mesh Ewald Continuous Constant pH Molecular Dynamics in Amber

Statistical machine learning for comparative protein dynamics with the DROIDS/maxDemon software pipeline - ScienceDirect
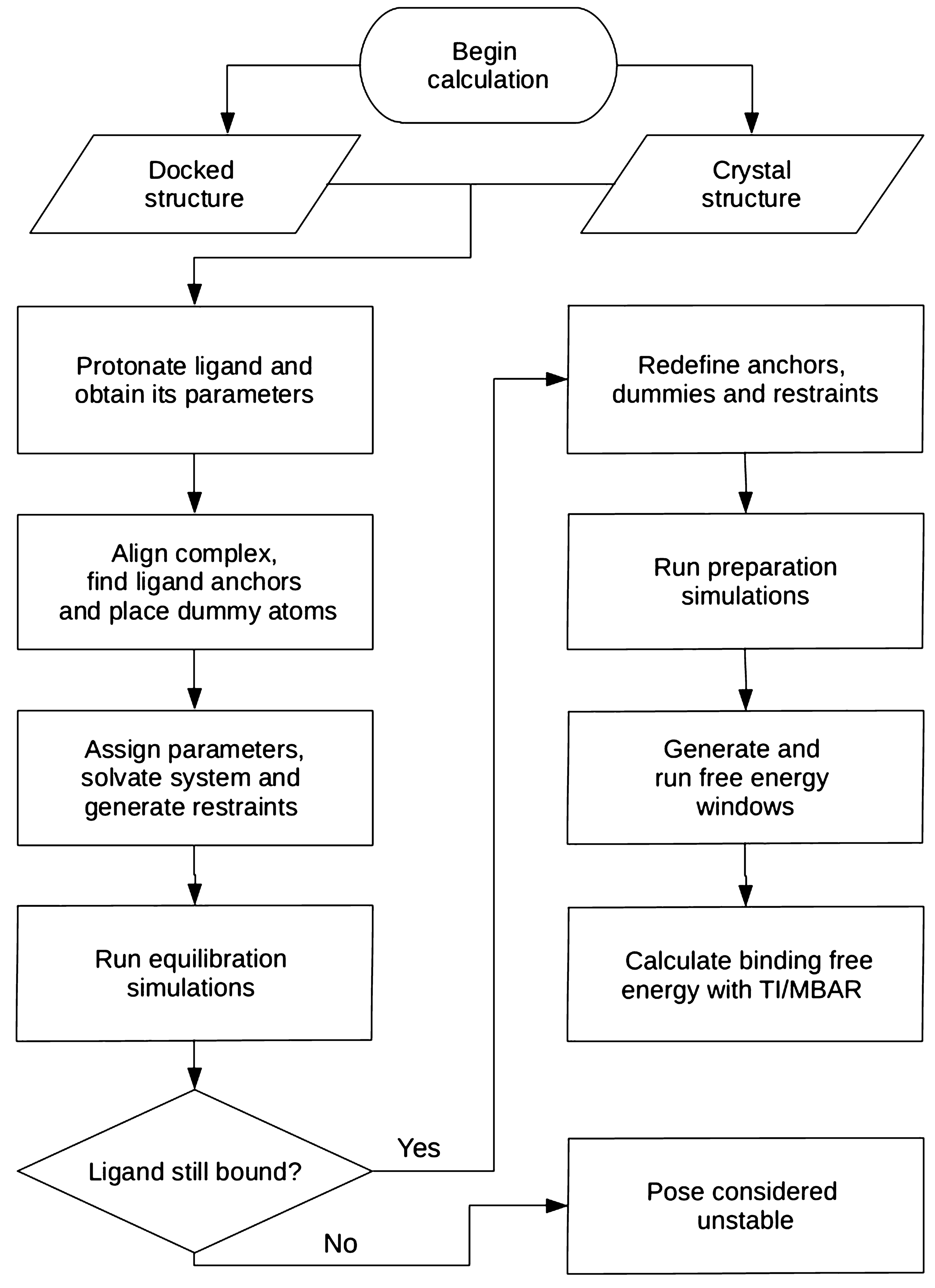
Automation of absolute protein-ligand binding free energy calculations for docking refinement and compound evaluation
de
por adulto (o preço varia de acordo com o tamanho do grupo)