Identification of levoglucosan degradation pathways in bacteria and sequence similarity network analysis
Por um escritor misterioso
Descrição
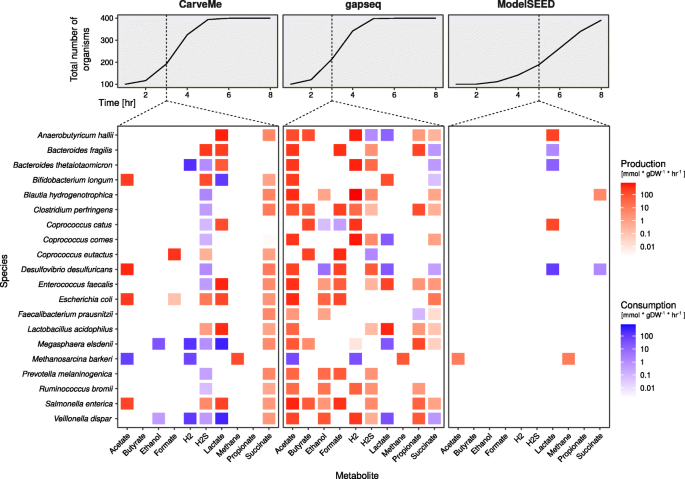
gapseq: informed prediction of bacterial metabolic pathways and reconstruction of accurate metabolic models, Genome Biology
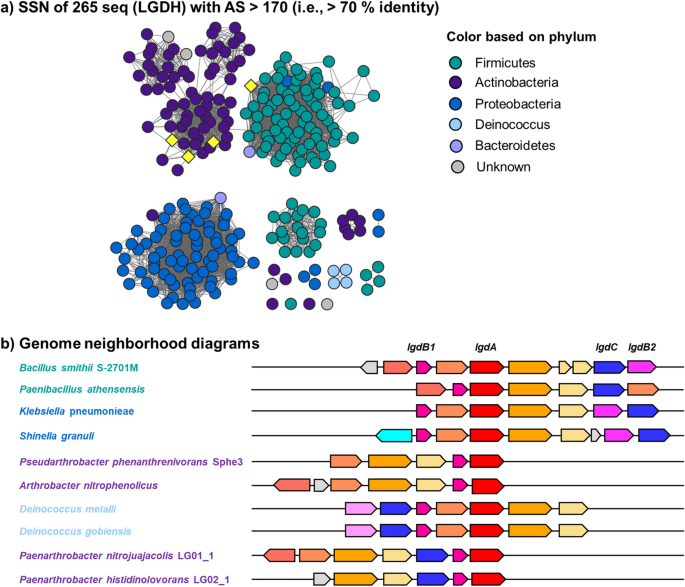
Identification of levoglucosan degradation pathways in bacteria and sequence similarity network analysis

Isolation and Characterization of Levoglucosan-Metabolizing Bacteria

Identification of levoglucosan degradation pathways in bacteria and sequence similarity network analysis
Domain shuffling in the enoyl-CoA hydratase family. The displayed
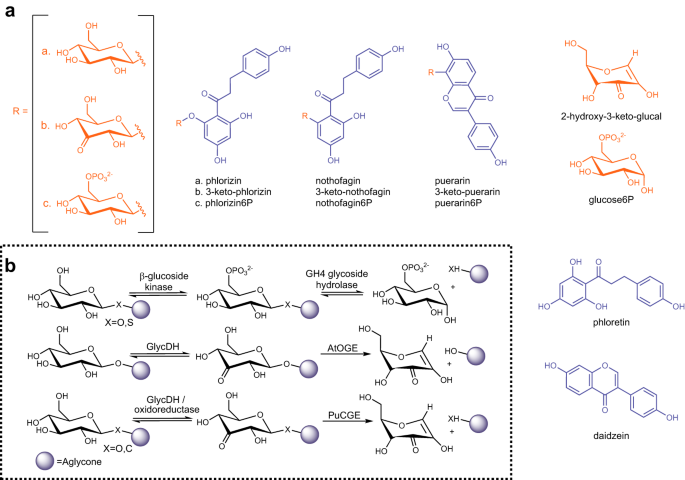
Enzymatic β-elimination in natural product O- and C-glycoside deglycosylation
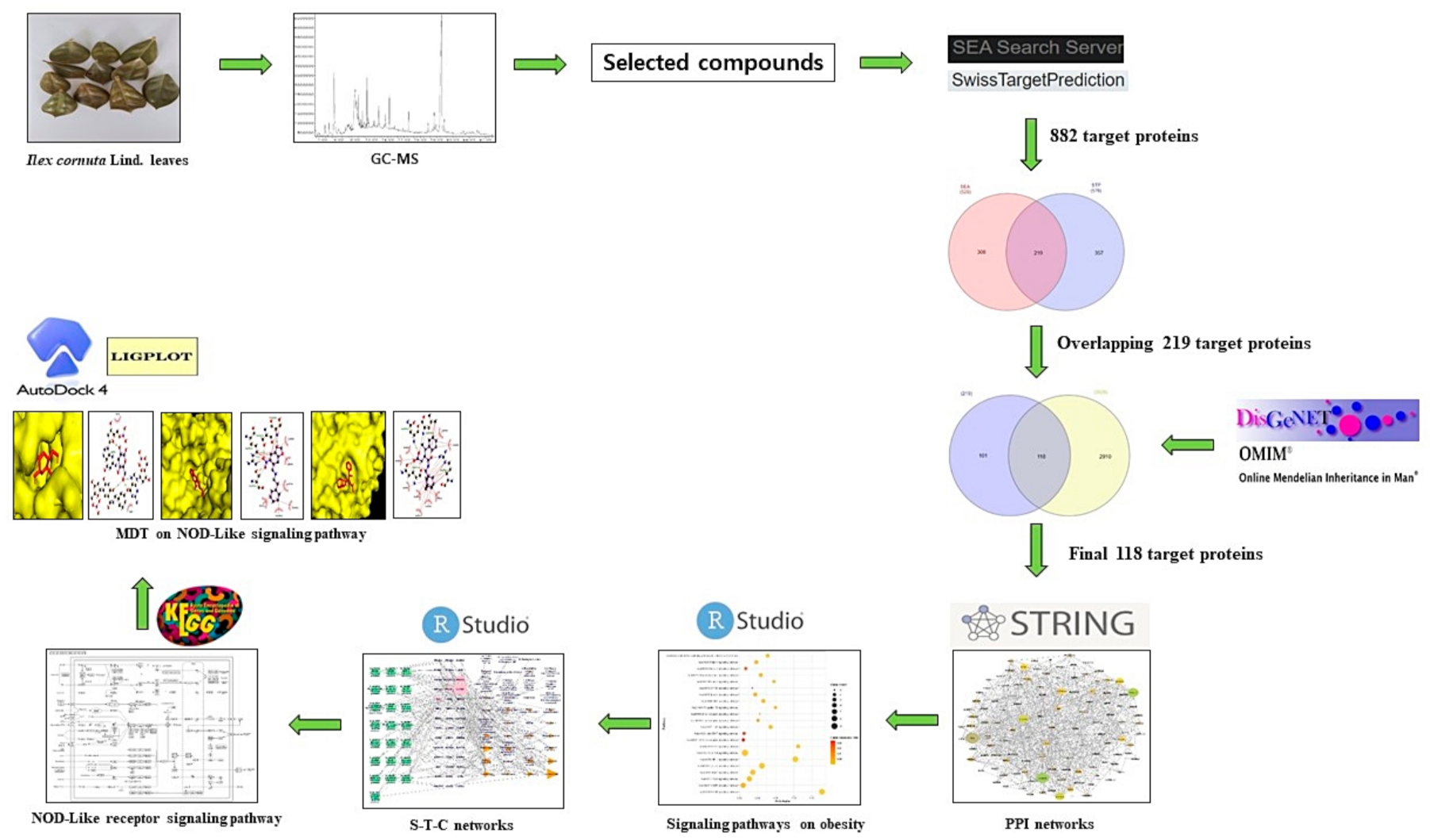
Processes, Free Full-Text
Publications - Scott lab
Identification of levoglucosan degradation pathways in bacteria and sequence similarity network analysis

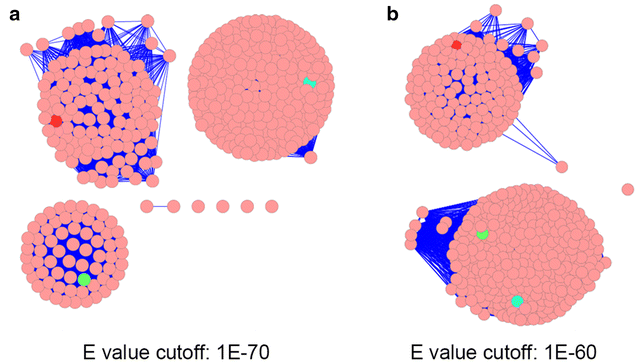
A. Thresholded sequence similarity networks represent sequences as
Publications - Scott lab
Sequence similarity network analysis, crystallization, and X-ray crystallographic analysis of the lactate metabolism regulator LldR from Pseudomonas aeruginosa, Bioresources and Bioprocessing
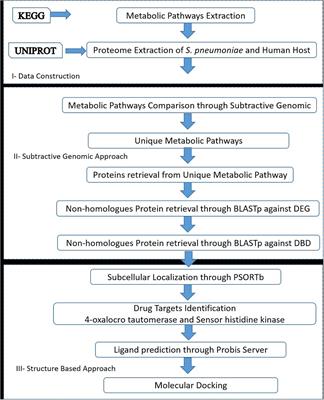
Frontiers Comparative Metabolic Pathways Analysis and Subtractive Genomics Profiling to Prioritize Potential Drug Targets Against Streptococcus pneumoniae

Production of levoglucosan (LG) by pyrolysis of cellulose, and
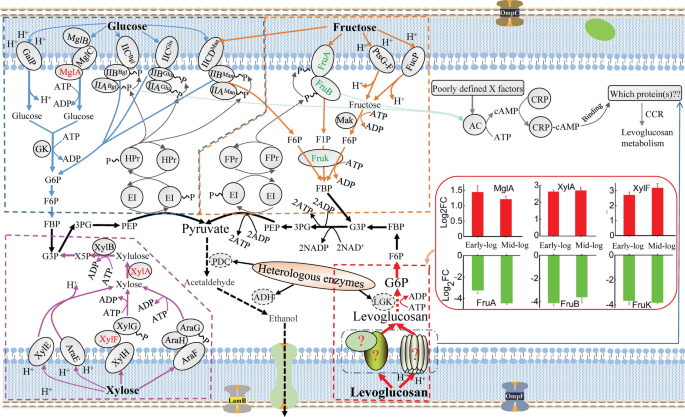
Omics analysis coupled with gene editing revealed potential transporters and regulators related to levoglucosan metabolism efficiency of the engineered Escherichia coli, Biotechnology for Biofuels and Bioproducts
de
por adulto (o preço varia de acordo com o tamanho do grupo)





:format(webp):quality(90)/https://www.playsport.ro/wp-content/uploads/2022/11/FC-Hermannstadt-CFR-Cluj.-Campioana-obligata-sa-castige-pe-terenul-revelatiei-turului-din-Superliga.jpg)